Abstract
Purpose
Exosomes are extracellular vesicles (EVs), 40-150 nm in diameter. They are released by all types of cells, and can potentially reflect the phenotypic state of the cell that produces them. Exosomes contain all the molecular constituents of a cell, including proteins, RNA, and DNA; they have been detected in many body fluids, including urine, cerebrospinal fluid, bile, ascites, and blood. Exosomes derived from chronic myelogenous leukemia (CML) cells may serve as novel targets for detecting the BCR-ABL mRNA transcript, and their contents may be used as a biomarker for the diagnosis and follow-up of patients with CML. However, no studies have been reported so far regarding this potential of exosomes. To explore the use of CML-derived exosomes as novel targets for detecting BCR-ABL mRNA transcripts, we isolated exosomes from two CML cell lines and from the sera of bone marrow (BM) blood of patients with CML, analyzed their characteristics, and performed PCR to measure the BCR-ABL mRNA transcript level.
Methods
Human CML cell lines (K562 and KU812) were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in an RPMI-1640 medium (Gibco, Carlsbad, CA, USA) with 10% exosome-depleted fetal bovine serum and 1% penicillin/streptomycin (Gibco). After culturing for 36-48 h, the supernatant was sequentially centrifuged at 5,000 × g for 30 min at 4 ºC, followed by another centrifugation at 10,000 × g for 30 min at 4 ºC to remove the detached cells. The exosomes were then isolated using a size-exclusion chromatography column packed with 10 mL of Sepharose CL-2B (GE Healthcare Life Sciences, Pittsburgh, PA, USA). The exosomes from the serum of BM blood were isolated with ExoQuickTM Exosome Precipitation Solution (SBI, Mountain View, CA, USA). Total RNA from whole cells and exosomes was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Next, 25-100 ng of purified RNA was reverse-transcribed using the Prime Script RT-PCR kit (TaKaRa BIO, Shiga, Japan). Nested PCR was performed to detect the BCR-ABL mRNA transcripts.
Results
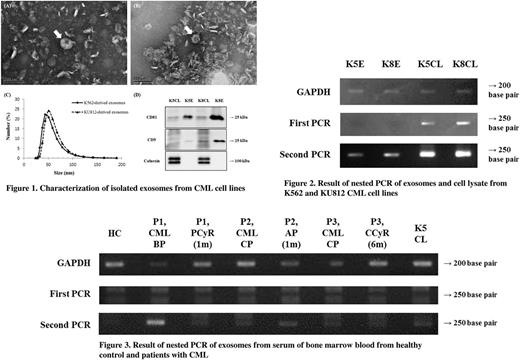
Transmission electron microscopy (TEM) was performed to study the morphology and size of the isolated exosomes. Exosomes derived from K562 and KU812 cells were found to be cup-shaped vesicles, 20-150 nm in diameter. Using dynamic light scattering, size distribution of the exosomes was found to be 30-190 nm in diameter. Exosome marker proteins (CD81 and CD9) were detected by western blotting; however, no endoplasmic reticulum marker protein (calnexin) was detected in the exosomes from either CML cell lines. This result confirmed that purified exosomes were obtained from the CML cell lines (Figure 1). Nested PCR was performed on these purified exosomes. A single band, corresponding to 250 bp, was detected in the exosomal RNA isolated from both the CML cell lines. RNA sequence analysis using NCBI/BLAST (National Center for Biotechnology Information, Bethesda, MD, USA) revealed 99% sequence homology between the detected 250-bp band and the partial mRNA for human BCR-ABL chimeric protein (Figure 2).
Nested PCR was also performed on exosomes isolated from the serum of BM blood (one healthy control and three patients with CML). Bands of about 250 bp were detected in the exosomal RNA isolated from the sera of patients with CML. RNA sequence analysis revealed that exosomal RNA from patients with CML at the blast phase and accelerated phase showed 99% sequence homology with the partial mRNA for human BCR-ABL chimeric protein. Bands were observed in exosomal RNA from patients with CML at the chronic phase as well, although not to the extent that RNA sequence analysis could proceed (Figure 3).
Conclusions
We found that exosomes isolated from CML cells and the sera of BM blood contained the BCR-ABL mRNA transcript. This study revealed that CML-derived exosomes may act as novel targets for the detection of the BCR-ABL mRNA transcript, and can be used as a biomarker.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal